
Module 5 – Genomics
Thursday, 2 December 2021
A genotype is just a genetic map, which is made up of traits which sets a plant or animal’s genetic potential.
In Newsletter 5 of the Genetics Technical Module we’re focusing on Genomics which underpins the accuracies of PIC’s genetic selections
Introduction
A genotype is just a genetic map, which is made up of traits which sets a plant or animal’s genetic potential. Each cell in a pig’s body has a nucleus containing the chromosomes which have that animal’s genetic information (Figure 1). Pigs have 19 chromosome pairs for a total of 38 chromosomes. Each chromosome contains many genes and every pig has two copies of each gene, one inherited from the sire, and one from the dam.
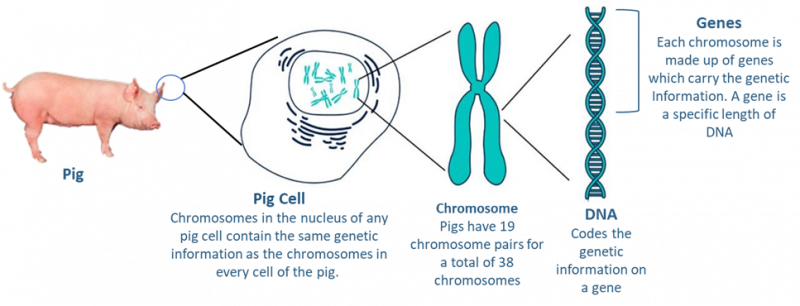
(Adapted from: https://www.bestfunquiz.com/q/how-much-you- know-about-chromosomes-and-genes)
While selecting elite animals should seem straightforward, it is more complex than just picking the best of the bunch. As technology has improved and our knowledge has increased, we’ve come to the realization that there is still a great deal to learn. Even though each pig’s cell only contains 38 chromosomes, each chromosome contains hundreds to thousands of genes, which in turn affect the traits or characteristics of the animal. In pig breeding we are interested in certain traits and what makes animal breeding and selection complex is that not only is each trait often regulated by more than one gene, some genes affect multiple and also unrelated traits. Over time, breeding programs have evolved from being very simple to very technologically complex.
The Evolution of Breeding Programs
The earliest breeding program were based on phenotype traits, the measurement of which used to select animals objectively i.e. to breed the best to the best. More recently, phenotype traits were used to estimate breeding values which were incorporated into indexes. This worked well but does have some downsides including:
- Genetic improvement in traits with low heritability can be limited and slow
- Some traits can only be measured properly after a long while or several reproductive cycles e.g. sow lifetime productivity
- Measurement of some traits can only be completed after the animal is processed e.g. Meat quality
- Several traits may be too costly to measure
- The emphasis on relatives may lead to greater rates of inbreeding
The next step up from phenotype-based selection was known as the Best Linear Unbiased Prediction or BLUP. BLUP is the most widespread system used to estimate Estimated Breeding Values (EBVs- Australian Pig Improver-Genetics Technical Module-2) Genetic Improvement Part A) and rank selected animals on genetic merit. It usesall available information to calculate EBVs. With BLUP, EBVs for selected animals are calculated using the phenotype information of these animals and all their relatives.
BLUP also corrects phenotypes for expected environmental effects, meaning it uses information for genetically related animals while also allowing for the fact that they may be located at different farms.
Marker Assisted Selection (MAS) followed and enhanced BLUP. This is a technique that uses DNA markers that associated with desirable traits to select animals or plants earlier on into a breeding program. MAS was first used in the pig industry in the early 1990s for marker assisted selection against the HAL gene which caused poor meat quality and also death in some stressful situations.
But MAS did not result in widespread use of DNA information in animal breeding as there are only a limited number of validated markers available, and these markers don’t explain enough of the genetic variation for each trait. The important traits in livestock turned out to be much more complicated than people thought as they can be impacted by single, multiple, sometimes thousands of genes that all had an effect on the animal’s phenotype.
Genomic selection on the other hand assumes a specific trait is affected all markers linked to a gene and focuses on figuring out their effect rather than their significance.
Genomic selection in pig breeding
The Genome of any living thing is the complete set of genetic material that is present in virtually every cell. Pig Genomics is the study of a pig’s genes (its genome), the interactions between these genes and the environment. Knowing the make-up of a pig’s genome, how it has evolved over time, and which genes impact on the different traits provide substantial information on the development and functioning of a pig.
Genomics has revolutionized the way that PIC is able to select elite animals in the Nucleus. By combining selection tools like BLUP with genomic information, PIC can accelerate genetic improvement by 30-35%.
Single Step Genomic BLUP (SSBLUP) which combines genomic and BLUP (phenotypic and pedigree) information has been shown to be 30-50% more accurate than BLUP based EBVs.
These higher accuracies provide the accelerated selection improvement with the greater knowledge provided by genomics, central to increased accuracies. For example, without genomic data, you don’t know the genetic make-up of the offspring of elite sows and boars; previously you assumed they were the same i.e. the “relatedness “of the littermates was 50% (Figure 2).
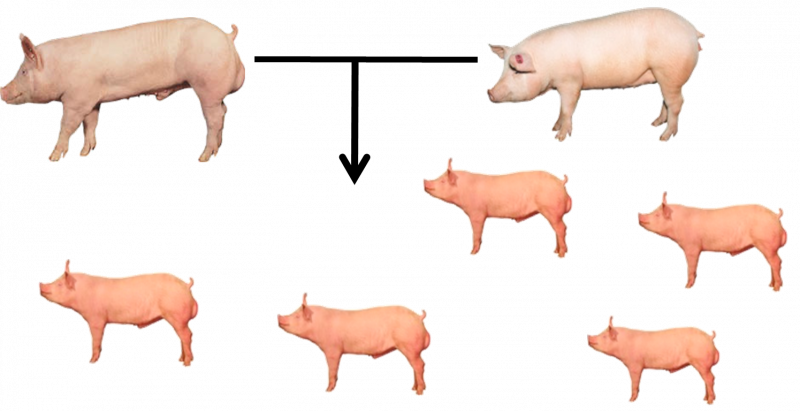
But what really happens is that the sow and boar each have a different genome, and from these two copies of the genome, different combinations of genes flagged by genetic markers are passed onto their piglets (Figure 3).
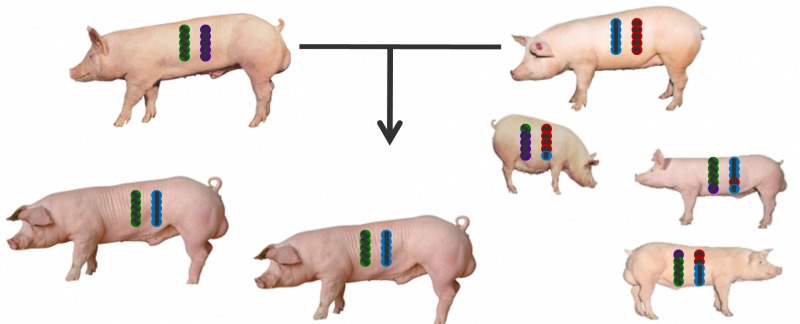
The genotypes determine what proportion of the genome the animals share (Figure 4).
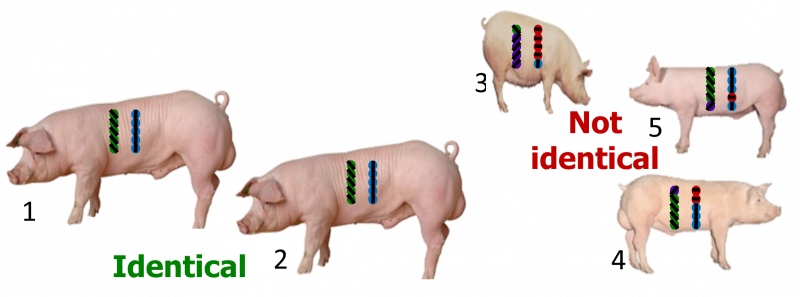
| Relationship | Littermates 1 & 2 | Littermate 3 | Littermate 4 | Littermate 5 |
| Assumed | 50% | 50% | 50% | 50% |
| True | 100% | 18% | 41% | 62% |
By using this innovative technology, PIC can produce the most scientifically accurate EBVs for the animals we select. This technology is also supported by substantial amounts of trait measurement data which is collected within our nucleus, multiplier, production and our client’s herds. This multi-disciplined approach and the use of cutting-edge technology provides a robust platform for ensuring selection accuracy for the PIC client.
References
The Genetics of the Pig. 2nd Edition. Edited by: Max Rothschild, Iowa State University, USA, Anatoly Ruvinsky, University of New England, NSW Australia. January 2011.
https://academic.oup.com/af/article/6/1/6/4638797
http://blog.biosearchtech.com/genomic-selection-can-it-accelerate-livestock-breeding
https://porkgateway.org/resource/basic-concepts-of-genetic-improvement/
https://thepigsite.com/articles/accuracies-of-prediction-in-pig-breeding
https://www.frontiersin.org/articles/10.3389/fgene.2019.00327/full
https://www.genome.gov/about-genomics/fact-sheets/A-Brief-Guide-to-Genomics
https://www.nationalhogfarmer.com/reproduction/blueprint-genome-enabled-selection-improves-pig-production
The opinions, advice and information contained in this publication are offered by PIC Australia soley for informational purposes and it does not constitute professional, commercial or technical advice. While information contained in this publication has been formulated in good faith, the contents do not take account all the factors which need to be considered before putting the information into practice. Accordingly, no person should rely on anything contained herein as a substitute for specific advice.

